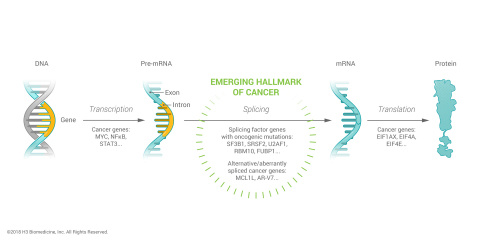
CAMBRIDGE, Mass.--(BUSINESS WIRE)--H3 Biomedicine Inc., a clinical stage biopharmaceutical company specializing in the discovery and development of next-generation cancer medicines using its data science and precision chemistry product engine, announced today the publication of a comprehensive genomic landscape analysis illuminating the breadth, frequency and potential disease-driving significance of somatic mutations in RNA splicing factor genes in human cancers. The findings demonstrate that splicing factor mutations, which lead to RNA splicing dysregulation, are highly prevalent in many hematologic and solid tumor cancers, suggestive of their role as a hallmark of tumor formation and growth and, thus, opportunities for therapeutic intervention. RNA splicing is the biological process by which pre-cursor messenger RNA (pre-mRNA) is edited into a mature messenger RNA (mRNA). Splicing factors carry out the editing process which is catalyzed by the core spliceosome complex. H3 Biomedicine scientists’ research and findings were published in the most recent online version of Cell Reports. (Seiler M. et al, “Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types”). This manuscript is part of The Cancer Genome Atlas (TCGA) Program, a joint effort of the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI).
“In recent years, somatic mutations in core splicing factors have been described in certain hematologic malignancies but the breadth of splicing factor mutations has been largely underappreciated. The findings published today give us for the first time a comprehensive understanding of just how common splicing factor mutations are across many types of cancer and of the role they may play in promoting tumor formation and progression,” said Markus Warmuth, M.D., President and Chief Executive Officer of H3 Biomedicine Inc. “This knowledge further validates the therapeutic approach of modulating spliceosome dysregulation to control tumor formation and growth, one of our core focus areas at H3 Biomedicine as evidenced by H3B-8800, our small molecule modulator of the SF3b complex, which is currently being evaluated in a Phase 1 clinical trial for patients with hematologic cancers and splicing factor mutations. In addition, the research findings open up an expansive, actionable trove of spliceosome targets that could be explored with novel therapeutic strategies.”
Using whole exome and RNA sequencing data in TCGA, H3 data scientists analyzed over 10,000 patient samples across 33 tumor types to identify somatic mutations in RNA splicing factors. The analysis of over 400 splicing factor genes identified 119 genes as having putative driver mutations, including over-representation, recurrent loss-of-function or recurrent hotspot (oncogene-like) mutations.
Mutations in splicing factor genes were found in all 33 tumor types with the frequency of mutations in multiple splicing factor genes as high as 60% in patient samples of the same tumor types, representing a much larger occurrence than previously reported. In addition, H3 scientists, as exemplified in case studies of five genes, showed that mutations in splicing factor genes led to dysregulated or altered splicing in mutated cells.
“This research and analysis exemplifies our differentiated drug discovery approach. We have the computational power and know-how to mine vast amounts of genomic data and reveal critical insights that we in turn utilize to develop and investigate highly targeted, precision approaches to cancer,” said Lihua Yu, Ph.D., Chief Data Science Officer of H3 Biomedicine Inc. “We’re particularly proud of this work because, beyond informing our own drug discovery efforts, it will help advance the entire field’s understanding of the role dysregulated RNA splicing plays in cancer.”
H3 Biomedicine is advancing novel cancer therapies that target core splicing factor mutations. A Phase 1 trial is underway in patients with hematologic malignancies for the company’s first spliceosome pathway-targeting cancer therapeutic, H3B-8800, a potent, selective and orally bioavailable small molecule modulator of wild-type and mutant SF3b complex, a splicing factor gene. The study is evaluating the safety and preliminary efficacy of H3B-8800 in patients with myelodysplastic syndromes, acute myeloid leukemia, and chronic myelomonocytic leukemia who carry mutations in splicing factor genes. Preclinical data demonstrate that H3B-8800 modulates RNA splicing and shows preferential antitumor activity in a range of spliceosome-mutant cancer models.
Splicing modulation is one of several research focus areas of H3 Biomedicine. H3 Biomedicine has two additional investigational therapies in Phase 1 clinical trials, including:
- H3B-6545, an oral, first-in-class ESR1 covalent antagonist targeting wild-type and mutant estrogen receptor α in endocrine-therapy resistant metastatic breast cancer patients; and
- H3B-6527, an oral, potent and highly selective small molecule covalent inhibitor of FGFR4 for treatment of hepatocellular carcinoma (HCC) patients with overexpression of FGF19.
About H3 Biomedicine Inc.
H3 Biomedicine, a Cambridge,
Massachusetts-based biopharmaceutical company specializing in the
discovery and development of precision oncology treatments, was
established in 2011 as a subsidiary of Eisai’s U.S. pharmaceutical
operation, Eisai Inc. H3 Biomedicine combines long-term vision with
operational excellence, leveraging its collaboration with Eisai Co.,
Ltd., who provides essential research funding and access to the
capabilities and resources of a global pharmaceutical company. Using
modern synthetic chemistry, chemical biology and human genetics, H3
Biomedicine seeks to bring the next generation of genomics-based cancer
treatments to market with the goal of improving the lives of patients.
For more information, please visit www.h3biomedicine.com.